OpenWorm: Building the First Comprehensive Computational Model of Caenorhabditis elegans
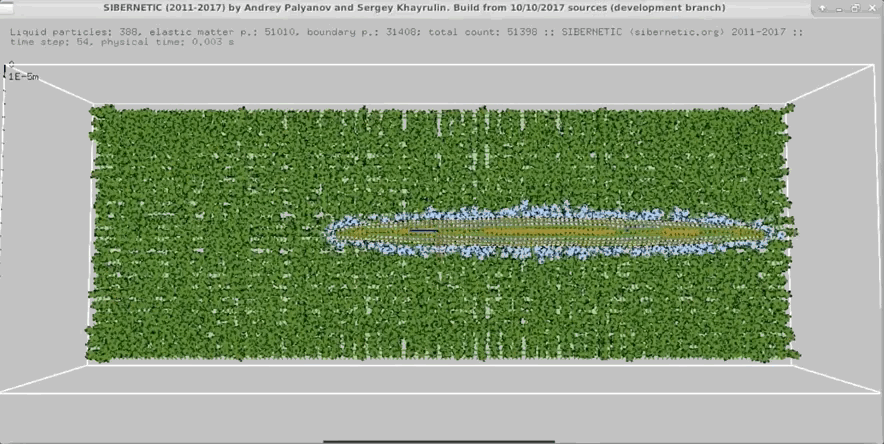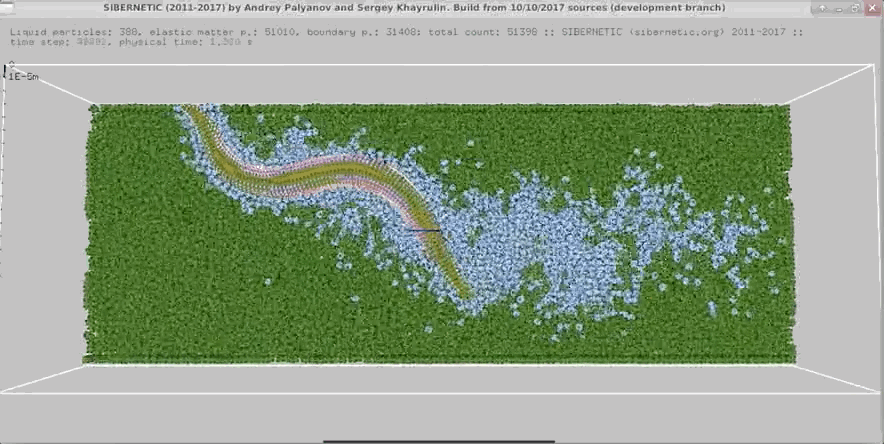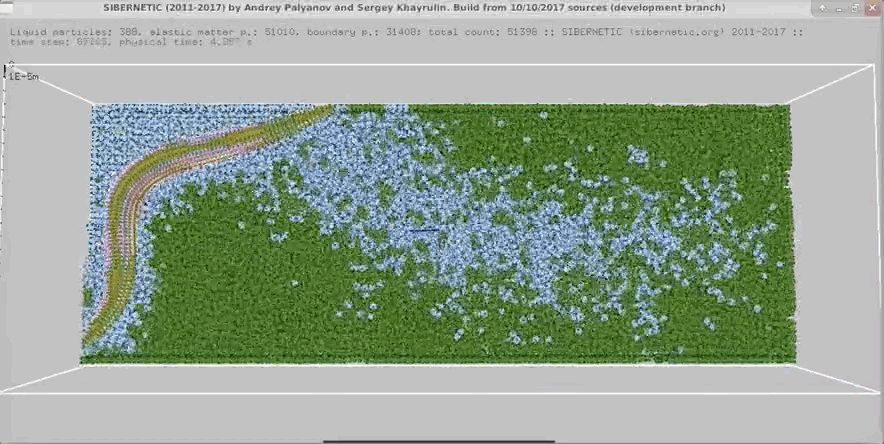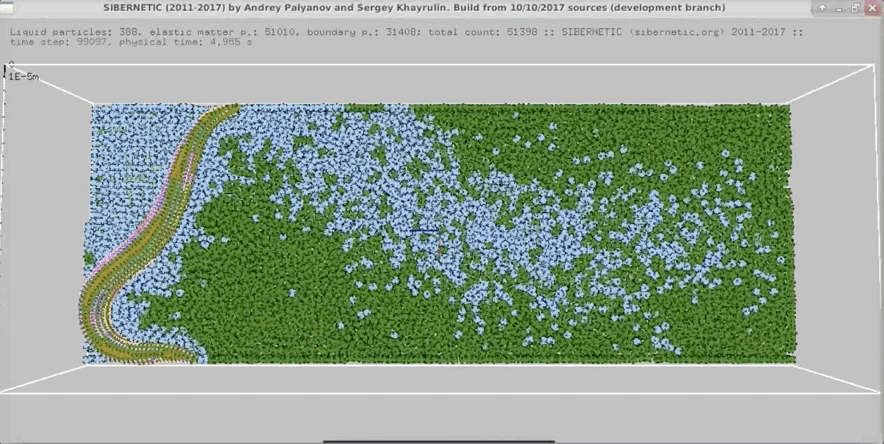
Imagine a tiny worm that is both real and entirely simulated. This mind-blowing concept raises questions about simulation theory and our understanding of reality. OpenWorm is an open-source project that aims to build the first comprehensive computational model of Caenorhabditis elegans (C. elegans), a microscopic roundworm. This article will break down how this simulation works and how you can run it on your own computer. By the end, you might be questioning your own reality!
What is OpenWorm?
OpenWorm is an open-source project and open science community dedicated to creating the world's first whole organism in a computer. The project focuses on C. elegans, a nematode, by using bottom-up systems biology and computational modeling. The main goal is to build a virtual organism to understand the events and mechanisms of living cells.
This groundbreaking project integrates data from scientific experiments carried out over the past decade. By incorporating available data from the scientific community into software models, OpenWorm can simulate the behavior of C. elegans. The project also collaborates with universities and research institutes to gather additional data and fill in gaps in existing knowledge.
The Worm
Caenorhabditis elegans is a small, transparent nematode about 1 mm in length that lives in soil, particularly in temperate regions. It's popular in biological research due to its simple properties. With a lifespan of about 2-3 weeks and 302 neurons, C. elegans is the first multicellular organism to have its entire genome sequenced. OpenWorm has mapped every single neuron and can predict the worm's reactions in any environment.
This level of detail allows researchers to simulate the worm's nervous system and observe its behavior in various conditions. By understanding how C. elegans functions on a cellular level, scientists can gain insights into more complex organisms, including humans.
Simulation Theory
Simulation theory suggests that we might be living in a computer simulation. Oxford philosopher Nick Bostrom proposed that humans could be living in a simulation created by an advanced civilization. With technology advancing rapidly, particularly in artificial intelligence and virtual reality, the possibility of creating realistic simulations is increasing. If we can simulate a simple organism like C. elegans, it raises the question of whether we could eventually simulate humans and the entire universe.
Nick Bostrom's theory posits that if it is possible to create simulations that are indistinguishable from reality, and if civilizations are interested in running such simulations, then it is highly likely that we are living in one. The rapid advancement of technology, including quantum computing, supports the plausibility of this hypothesis. Imagine the compute power needed to simulate humans with 86 billion neurons, far surpassing the 302 neurons in C. elegans.
Using Docker for OpenWorm Simulations
Docker is a platform that allows you to run software in isolated environments called containers. OpenWorm uses Docker to run its simulations, making it easier for anyone to set up and run the worm simulation on their own computer. This setup ensures consistency across different systems and simplifies the installation process.
Benefits of Using Docker:
- Isolation: Docker containers isolate the simulation environment from the host system, ensuring that dependencies and configurations do not conflict.
- Portability: Docker images can be run on any system that supports Docker, making it easy to share and collaborate.
- Reproducibility: Using Docker ensures that the simulation runs the same way every time, which is crucial for scientific research.
Step-by-Step Guide to Running OpenWorm
Prerequisites for Installation
Before installing OpenWorm, ensure your system meets the following requirements:
- At least 60 GB of free space
- At least 2 GB of RAM
- Ability to clone git repositories (install git if necessary)
Installation Instructions
- Install Docker: Download and install Docker from Docker's website according to your operating system.
- Clone the OpenWorm Repository: Open a terminal and run:
git clone http://github.com/openworm/openworm
- Navigate to the OpenWorm Directory: Change to the OpenWorm directory:
cd openworm
- Optional - Build the Docker Image: This step builds the Docker image locally:
./build.sh
For Windows, use:
build.cmd
If you skip this step, the latest released Docker image will be downloaded automatically.
- Run the Simulation: Execute the following command to start the simulation:
./run.sh
For Windows, use:
run.cmd
- Monitor the Simulation Output: The simulation will produce output over 5-10 minutes, displaying on the terminal screen.
- Clean Up: After the simulation ends, clean up the Docker container by running:
./stop.sh
For Windows, use:
stop.cmd
- Inspect the Output: Check the
outputdirectory on your local machine for the simulation results.
Example Outputs from OpenWorm Simulations
Running the OpenWorm simulation produces various outputs that help researchers analyze the worm's behavior and nervous system activity. These outputs include graphs, visualizations, and 3D animations.
Graphs and Visualizations
The nervous system model, c302, generates detailed graphs showing the activity of neurons. These graphs help researchers understand how neurons interact and respond to different stimuli. The 3D body model, Sibernetic, produces visualizations that illustrate the worm's movement and muscle activity. These visualizations provide insights into the biomechanical properties of C. elegans.
3D Body Model Animations
The simulation also creates 3D animations of the worm's movements. These animations show how the worm crawls, interacts with its environment, and performs various behaviors. By examining these animations, researchers can validate the accuracy of the simulation and make necessary adjustments to improve it.
Analyzing the Output Data
The data produced by the simulation is stored in the output directory. This data includes neuron activity logs, muscle activity logs, and other relevant information. Researchers can analyze this data to gain a deeper understanding of the worm's behavior and the underlying biological processes.
Advanced Usage and Customization
OpenWorm offers several advanced features and customization options for users who want to delve deeper into the simulation and modify it according to their needs.
Modifying Simulation Duration
The duration of the simulation can be adjusted using the -d argument. The default duration is 15 milliseconds, but you can extend it to observe more detailed behavior. For example, to run the simulation for 5000 milliseconds (5 seconds), use the following command:
./run.sh -d 5000
For Windows, use:
run.cmd -d 5000
Exploring Docker Container Internals
You can explore the internals of the Docker container by running a special script that logs you into the container before it executes the main simulation script. This allows you to inspect and modify the installed systems and code bases. Use the following command:
./run-shell-only.sh
For Windows, use:
run-shell-only.cmd
Once inside the container, you can use terminal editors like nano to modify the code. After making changes, remember to clean up by running stop.sh.
Customizing Installations and Running Processes
To customize what gets installed in the Docker container, modify the Dockerfile. To change what runs during the simulation, edit the master_openworm.py script. After making these changes, rebuild the Docker image with the following command:
./build.sh
For Windows, use:
build.cmd
Then, run the simulation as usual.
Troubleshooting and FAQs
OpenWorm provides solutions to common issues and answers frequently asked questions to help users troubleshoot problems and optimize their simulation experience.
Common Issues and Their Solutions
- Insufficient Disk Space: Ensure you have at least 60 GB of free space. If using an external hard disk, specify the location for image storage in Docker preferences.
- Docker Daemon Not Running: Make sure the Docker daemon is running in the background. Look for the Docker whale logo in the menu bar or system tray.
Accessing and Extracting Data from Docker
To access additional data generated by the simulation, use the docker cp command to copy files from the Docker container to your local machine. For example, to copy the worm motion log:
docker cp openworm:/home/ow/sibernetic/simulations/[SPECIFIC_TIMESTAMPED_DIRECTORY]/worm_motion_log.txt ./worm_motion_log.txt
Replace [SPECIFIC_TIMESTAMPED_DIRECTORY] with the appropriate directory name.
Understanding Docker Container Commands
exit: Stops execution in the Docker container and returns to the system shell.stop.sh: Removes the container and associated files, resetting it for a new run.- Re-entering the Docker Container: Use the following commands to re-enter an exited container:
docker start openworm
docker exec -it openworm /bin/bash
Community and Collaboration
OpenWorm thrives on community contributions and collaborations with universities and research institutes. By joining the OpenWorm community, you can participate in groundbreaking research, share ideas, and collaborate with scientists worldwide.
OpenWorm Community Contributions
The OpenWorm project is supported by a global community of volunteers who contribute their expertise in biology, neuroscience, computer science, and other fields. These contributors help develop and refine the models, conduct experiments, and analyze data.
Collaborations with Universities and Research Institutes
OpenWorm collaborates with several prestigious institutions, including:
- Salk Institute: Data and assistance from Dr. Sreekanth Chalasani.
- UMass Medical School: Contributions from Dr. Michael Francis.
- University of Cambridge: Support from Dr. William Schafer.
- Princeton University: Collaboration with Dr. Andrew Leifer.
- Leeds University: Worm simulations guided by Dr. Netta Cohen and her students.
- Caltech: 3D images of the worm’s body provided by the Virtual Worm project.
How to Get Involved with OpenWorm
Joining the OpenWorm community is easy. Visit the OpenWorm website and explore the various ways you can contribute. Whether you're a biologist, programmer, or enthusiast, your involvement can make a significant impact.
Documentation and Further Resources
OpenWorm provides extensive documentation and resources to support the research community and facilitate collaboration. These resources include comprehensive guides, scientific data, and interactive tools.
Comprehensive Documentation Overview
The OpenWorm documentation site contains all the necessary information to get started with the project. It includes installation guides, simulation instructions, and detailed descriptions of the models and tools used in the project.
Joining OpenWorm Slack Community
Connect with other OpenWorm contributors and stay updated on the latest developments by joining the OpenWorm Slack community. Get an invite here.
High-Level Issues and Milestones
Track the project's progress and contribute to discussions on high-level issues and milestones by visiting the OpenWorm GitHub repository. Here, you can find and report bugs, suggest new features, and collaborate on solutions.
Earning OpenWorm Badges
OpenWorm offers badges to recognize the contributions and achievements of its community members. By participating in the project and completing specific tasks, you can earn these badges.
Introduction to OpenWorm Badges
Badges are awarded for various contributions, such as running simulations, developing new features, and helping with documentation. These badges serve as a form of recognition and can be displayed on your profile.
Steps to Earn a Docker Badge
To earn the Docker badge, follow these steps:
- Set up and run the OpenWorm Docker container on your computer.
- Complete the simulation and analyze the output.
- Submit your results and contributions to the OpenWorm community for review.
Importance of Community Recognition
Earning badges not only recognizes your contributions but also helps build your reputation within the community. It encourages active participation and fosters a sense of achievement.
Case Studies and Success Stories
OpenWorm has achieved significant milestones, showcasing the power of collaborative research and computational modeling. These case studies highlight the project's impact and future potential.
Notable Achievements within the OpenWorm Project
Some of the key achievements include:
- First Virtual Organism: Successfully creating a comprehensive computational model of C. elegans.
- Detailed Neuron Mapping: Mapping all 302 neurons and their connections.
- Integration of Multiple Models: Combining nervous system and 3D body models to simulate realistic behavior.
Real-World Applications of OpenWorm Research
The research conducted by OpenWorm has applications in various fields, such as:
- Neuroscience: Understanding how neurons interact and process information.
- Computational Biology: Developing more accurate models of biological systems.
- Artificial Intelligence: Enhancing AI algorithms based on biological principles.
Future Directions and Potential Developments
OpenWorm continues to push the boundaries of computational biology. Future directions include:
- Expanding the Model: Adding more biological details and improving accuracy.
- Educational Tools: Developing interactive tools for teaching and learning biology.
- Collaborative Research: Engaging more researchers and institutions in the project.
Conclusion
OpenWorm is revolutionizing the field of computational biology by creating a comprehensive model of C. elegans. This project not only advances our understanding of this simple organism but also raises profound questions about simulation theory and the nature of reality. Join the OpenWorm community and contribute to this groundbreaking research.